Cryo-EM structures of Rho1 GABAA receptors with antagonist and agonist drugs
Fan, C., Cowgill, J., Howard, R.J., Lindahl, E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma-aminobutyric acid receptor subunit rho-1 | 479 | Homo sapiens | Mutation(s): 0 Gene Names: GABRR1 | 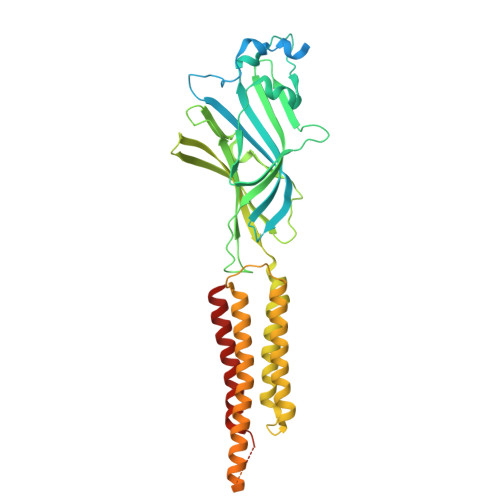 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P24046 (Homo sapiens) Explore P24046 Go to UniProtKB: P24046 | |||||
PHAROS: P24046 GTEx: ENSG00000146276 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P24046 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | AB [auth E] BB [auth E] DA [auth C] EA [auth C] G [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| D12 Query on D12 | BA [auth B], IB [auth E], LA [auth C], O [auth A], XA [auth D] | DODECANE C12 H26 SNRUBQQJIBEYMU-UHFFFAOYSA-N |  | ||
| D10 Query on D10 | AA [auth B] CA [auth C] HB [auth E] KA [auth C] N [auth A] | DECANE C10 H22 DIOQZVSQGTUSAI-UHFFFAOYSA-N |  | ||
| EI7 Query on EI7 | F [auth A], NA [auth D], P [auth A], S [auth B], YA [auth E] | 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one C6 H8 N2 O2 ZXRVKCBLGJOCEE-UHFFFAOYSA-N |  | ||
| OCT Query on OCT | FB [auth E], IA [auth C], L [auth A], UA [auth D], Y [auth B] | N-OCTANE C8 H18 TVMXDCGIABBOFY-UHFFFAOYSA-N |  | ||
| HEX Query on HEX | CB [auth E] DB [auth E] EB [auth E] FA [auth C] GA [auth C] | HEXANE C6 H14 VLKZOEOYAKHREP-UHFFFAOYSA-N |  | ||
| CL Query on CL | MA [auth C] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swedish Research Council | Sweden | 2019-02433 |