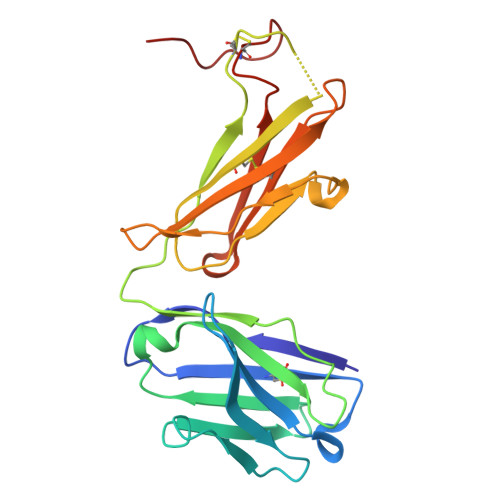
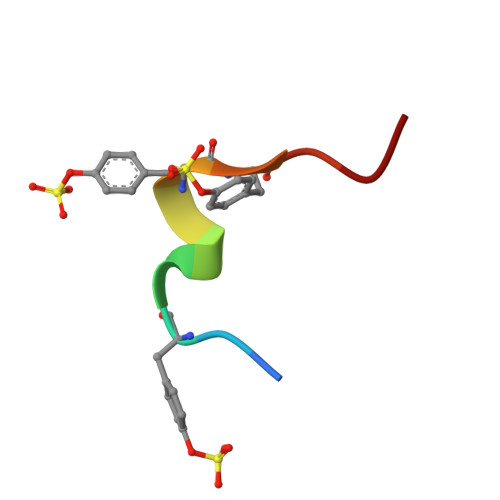
Specific recognition mechanism of an antibody to sulfated tyrosine and its potential use in biological research.
Ujiie, K., Nakakido, M., Kinoshita, S., Caaveiro, J.M.M., Entzminger, C.K., Okumura, S.C.J., Maruyama, T., Miyauchi, K., Matano, T., Tsumoto, K.(2025) J Biological Chem 301: 108176-108176
- PubMed: 39798874
- DOI: https://doi.org/10.1016/j.jbc.2025.108176
- Primary Citation of Related Structures:
9J8A - PubMed Abstract:
Post-translational modification of proteins is a crucial biological reaction that regulates protein functions by altering molecular properties. The specific detection of such modifications in proteins has made significant contributions to molecular biology research and holds potential for future drug development applications. In HIV research, for example, tyrosine sulfation at the N-terminus of C-C chemokine receptor type 5 (CCR5) is considered to significantly enhance HIV infection efficiency. However, antibodies specific to sulfated CCR5 still need to be developed. In this study, we successfully generated an antibody that specifically recognized the sulfated N-terminal peptide of CCR5 through rabbit immunization and panning via phage display using a CCR5 N-terminal peptide containing sulfate modification. We used various physicochemical methods in combination with molecular dynamics simulation to screen for residues that could be involved in recognition of the sulfated peptide by this antibody. We also confirmed that this antibody recognized the sulfated full-length CCR5 on the cell surface, which suggested it should be useful as a research tool that could lead to the development of novel therapeutics. Although the antibody binding did not inhibit HIV infection, it could be also described as sulfation site-specific binding, beyond sulfation-specific binding.
Organizational Affiliation:
Department of Bioengineering, School of Engineering, The University of Tokyo, Tokyo, Japan.